

The goal of regport is to provides R6 classes, methods and utilities to construct, analyze, summarize, and visualize regression models (CoxPH and GLMs).
You can install the development version of regport like so:
This is a basic example which shows you how to build and visualize a Cox model.
Prepare data:
Create a model:
model = REGModel$new(
lung,
recipe = list(
x = c("age", "sex"),
y = c("time", "status")
)
)
#> data.frame [228, 3]
#> survival::Surv(time, status) Surv 306 455 1010 210 883 1022~
#> age dbl 74 68 56 57 60 74
#> sex fct 1 1 1 1 1 1
model
#> <REGModel> ==========
#>
#> Parameter | Coefficient | SE | 95% CI | z | p
#> -----------------------------------------------------------------
#> age | 1.02 | 9.38e-03 | [1.00, 1.04] | 1.85 | 0.065
#> sex [2] | 0.60 | 0.10 | [0.43, 0.83] | -3.06 | 0.002
#>
#> Uncertainty intervals (equal-tailed) and p values (two-tailed) computed using a
#> Wald z-distribution approximation.
#> [coxph] model ==========You can also create it with formula:
model = REGModel$new(
lung,
recipe = Surv(time, status) ~ age + sex
)
#> data.frame [228, 3]
#> Surv(time, status) Surv 306 455 1010 210 883 1022 1 1 0 1 1~
#> age dbl 74 68 56 57 60 74
#> sex fct 1 1 1 1 1 1
model
#> <REGModel> ==========
#>
#> Parameter | Coefficient | SE | 95% CI | z | p
#> -----------------------------------------------------------------
#> age | 1.02 | 9.38e-03 | [1.00, 1.04] | 1.85 | 0.065
#> sex [2] | 0.60 | 0.10 | [0.43, 0.83] | -3.06 | 0.002
#>
#> Uncertainty intervals (equal-tailed) and p values (two-tailed) computed using a
#> Wald z-distribution approximation.
#> [coxph] model ==========Visualize it:
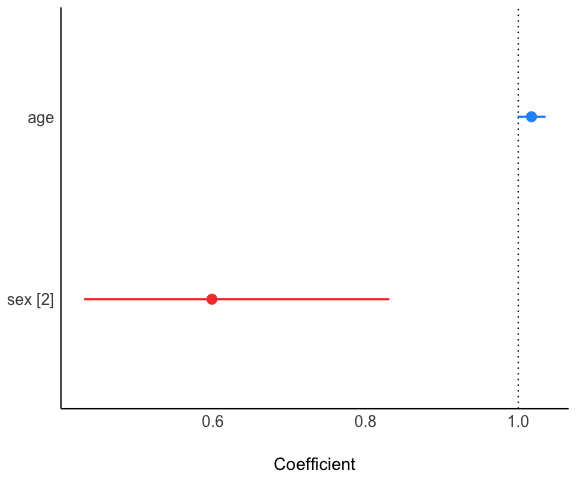
Visualize with more nice forest plot.
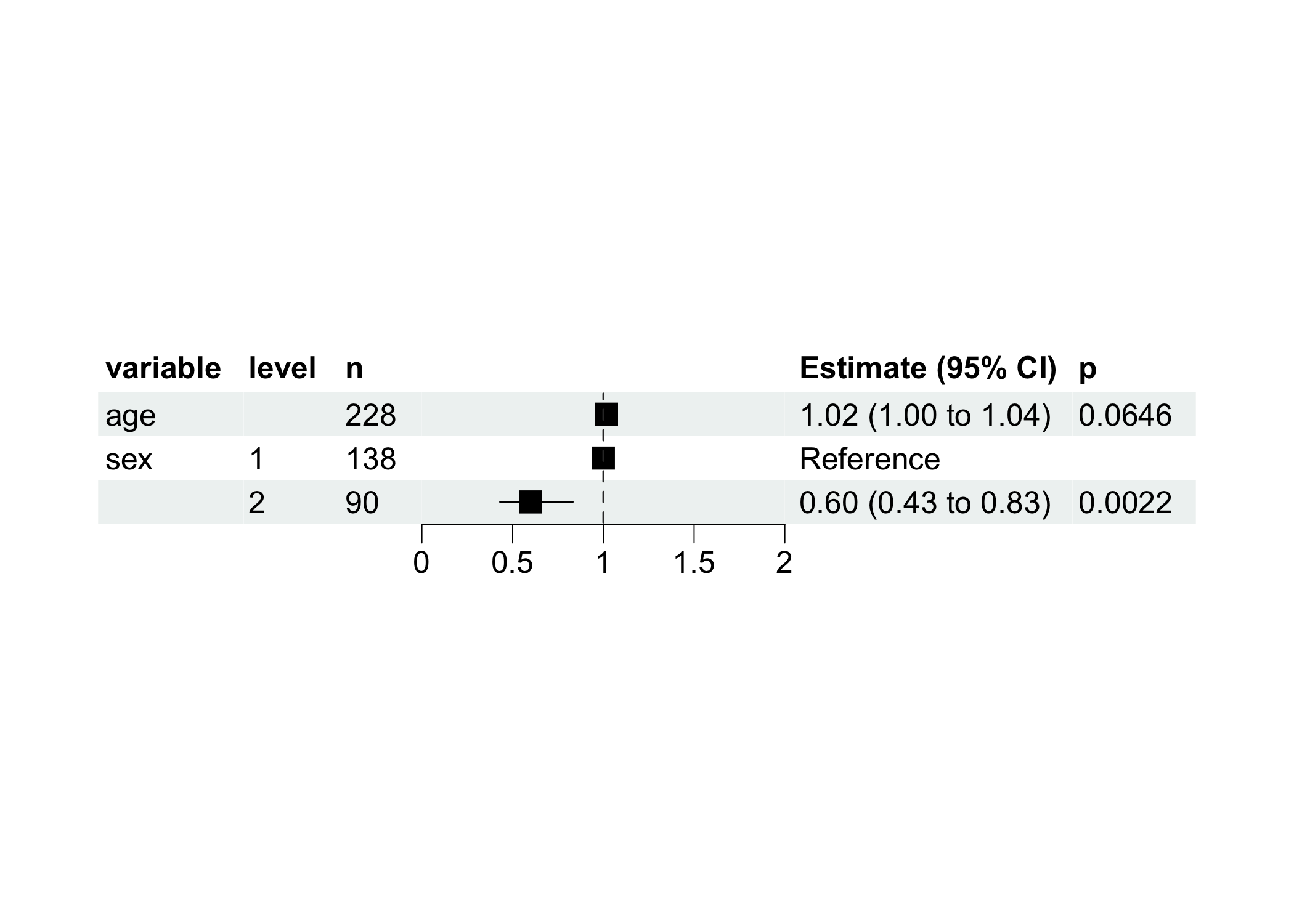
ml <- REGModelList$new(
data = mtcars,
y = "mpg",
x = c("factor(cyl)", colnames(mtcars)[3:5]),
covars = c(colnames(mtcars)[8:9], "factor(gear)")
)
ml
#> <REGModelList> ==========
#>
#> X(s): factor(cyl), disp, hp, drat
#> Y(s): mpg
#> covars: vs, am, factor(gear)
#>
#> Not build yet, run $build() method
#> [] model ==========
ml$build(f = "gaussian")
#> data.frame [32, 5]
#> mpg dbl 21 21 22.8 21.4 18.7 18.1
#> factor(cyl) fct 6 6 4 6 8 6
#> vs dbl 0 0 1 1 0 1
#> am dbl 1 1 1 0 0 0
#> factor(gear) fct 4 4 4 3 3 3
#> data.frame [32, 5]
#> mpg dbl 21 21 22.8 21.4 18.7 18.1
#> disp dbl 160 160 108 258 360 225
#> vs dbl 0 0 1 1 0 1
#> am dbl 1 1 1 0 0 0
#> factor(gear) fct 4 4 4 3 3 3
#> data.frame [32, 5]
#> mpg dbl 21 21 22.8 21.4 18.7 18.1
#> hp dbl 110 110 93 110 175 105
#> vs dbl 0 0 1 1 0 1
#> am dbl 1 1 1 0 0 0
#> factor(gear) fct 4 4 4 3 3 3
#> data.frame [32, 5]
#> mpg dbl 21 21 22.8 21.4 18.7 18.1
#> drat dbl 3.9 3.9 3.85 3.08 3.15 2.76
#> vs dbl 0 0 1 1 0 1
#> am dbl 1 1 1 0 0 0
#> factor(gear) fct 4 4 4 3 3 3
str(ml$result)
#> Classes 'data.table' and 'data.frame': 25 obs. of 10 variables:
#> $ focal_term: chr "factor(cyl)" "factor(cyl)" "factor(cyl)" "factor(cyl)" ...
#> $ variable : chr "(Intercept)" "factor(cyl)6" "factor(cyl)8" "vs" ...
#> $ estimate : num 23.28 -5.34 -8.5 1.68 4.31 ...
#> $ SE : num 3.1 1.89 3.05 2.35 2.16 ...
#> $ CI : num 0.95 0.95 0.95 0.95 0.95 0.95 0.95 0.95 0.95 0.95 ...
#> $ CI_low : num 17.203 -9.04 -14.473 -2.931 0.084 ...
#> $ CI_high : num 29.37 -1.64 -2.53 6.3 8.54 ...
#> $ t : num 7.504 -2.829 -2.791 0.715 1.999 ...
#> $ df_error : int 25 25 25 25 25 25 25 26 26 26 ...
#> $ p : num 6.18e-14 4.67e-03 5.25e-03 4.75e-01 4.56e-02 ...
#> - attr(*, ".internal.selfref")=<externalptr>
str(ml$forest_data)
#> Classes 'data.table' and 'data.frame': 6 obs. of 17 variables:
#> $ focal_term: chr "factor(cyl)" "factor(cyl)" "factor(cyl)" "disp" ...
#> $ variable : chr "factor(cyl)" NA NA "disp" ...
#> $ term : chr "factor(cyl)4" "factor(cyl)6" "factor(cyl)8" "disp" ...
#> $ term_label: chr "factor(cyl)" "factor(cyl)" "factor(cyl)" "disp" ...
#> $ class : chr "factor" "factor" "factor" "numeric" ...
#> $ level : chr "4" "6" "8" NA ...
#> $ level_no : int 1 2 3 NA NA NA
#> $ n : int 11 7 14 32 32 32
#> $ estimate : num 0 -5.3404 -8.5026 -0.0282 -0.0515 ...
#> $ SE : num NA 1.88767 3.04626 0.00924 0.01201 ...
#> $ CI : num NA 0.95 0.95 0.95 0.95 0.95
#> $ CI_low : num NA -9.0402 -14.4732 -0.0463 -0.075 ...
#> $ CI_high : num NA -1.6407 -2.532 -0.0101 -0.0279 ...
#> $ t : num NA -2.83 -2.79 -3.05 -4.28 ...
#> $ df_error : int NA 25 25 26 26 26
#> $ p : num NA 4.67e-03 5.25e-03 2.27e-03 1.84e-05 ...
#> $ reference : logi TRUE FALSE FALSE FALSE FALSE FALSE
#> - attr(*, ".internal.selfref")=<externalptr>
ml$plot_forest(ref_line = 0, xlim = c(-15, 8))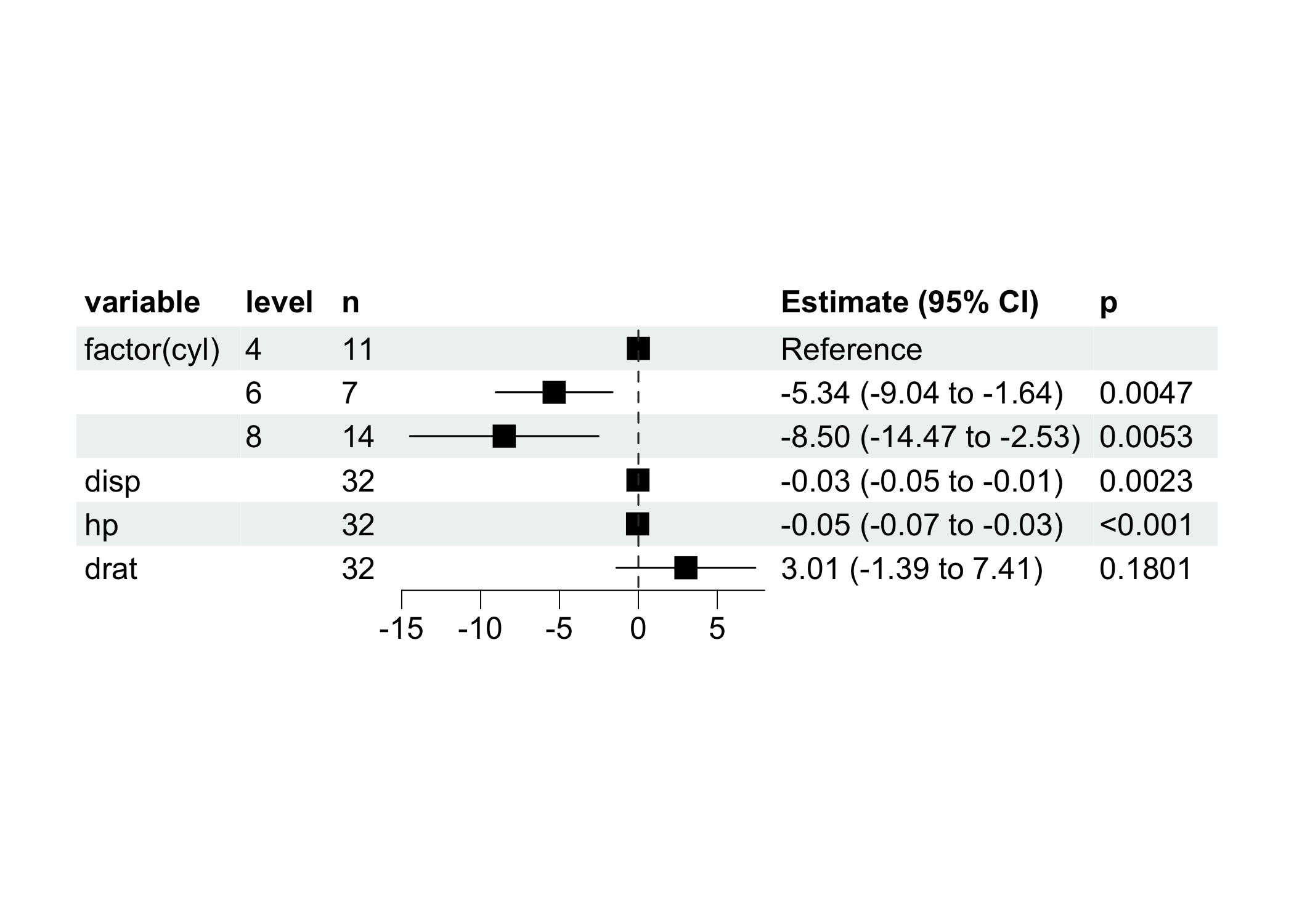
covr::package_coverage()
#> regport Coverage: 83.27%
#> R/utils.R: 75.00%
#> R/REGModelList.R: 79.31%
#> R/REGModel.R: 85.14%(MIT) Copyright (c) 2022 Shixiang Wang